The Host-Microbiome Center’s Molecular Unit manages molecular analyses of host, commensal, pathogenic microbial species. Internally, the core handles nucleic acid extraction of primary samples, and primary cultures, and high-throughput molecular analyses using multiplex PCR and next-generation-sequencing (NGS) platforms for 16SrRNA gene phylotyping and bacterial genome sequencing by Illumina MiSeq. The Center works with other area cores to provide HiSeq based sequencing for metagenomic and metatranscriptomic analyses. The core provides quality control and filtered sequencing datasets and suggests working with the Computational Unit if further analyses are needed.
The unit maintains high-throughput liquid handlers for nucleic acid extraction, quantification and library preparation, high-throughput single or multiplex qPCR/rtPCR studies, and NGS by Illumina MiSeq
The Molecular and Microbiology Units supports the multi-institutional Prospective Pathogen Genomic Surveillance program at Partners Healthcare and has also been instrumental in developing defined consortia of human commensal species for the treatment of food allergies and other diseases.
Example Projects:
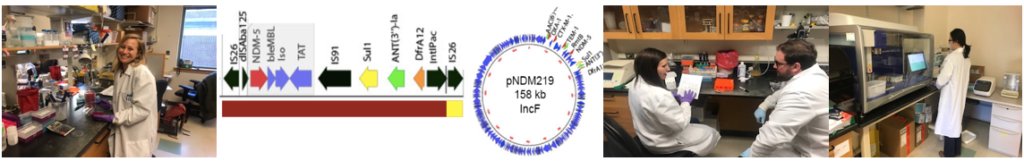
- Genome sequencing of carbapenem resistant Enterobacteriaceae (CRE) in a multi-hospital prospective genomic surveillance program with reporting to support local infection control teams.
- 16S rRNA gene phylotyping to evaluate longitudinal microbiome dynamics during infection and with dietary changes.
Case Studies:
Pathogen Genomic Surveillance Program
Molecular Analyses that support development of the MDSINE (Microbial Dynamical Systems Inference Engine for microbiome time-series analyses)
To inquire further please fill out our online request form or contact us at microbiome@bwh.harvard.edu.
